Clustering with matrix Factorization for anomaly detection
April 29, 2020Anomaly detection is a general problem that has been approached from several different fields. In most, but not all, cases anomaly detection is powered by some statistical property of the sample distribution. In a new paper [1], a method is presented based on clustering that perform outlier detection by sorting observations by the distance to clusterers. The method incorporates a similarity metric that drives the clustering algorithm to cluster samples together that are similar in a graph-traversing sense.
The clustering proceedure is based on non-negative matrix factorization (NMF) and supported by a graph-traversing algorithm called Minimum Spanning Tree. NMF has found great success in recommender systems but it is also applied in other fields such as astronomy, audio signal processing and other clustering tasks.
In this article:
- Non-negative matrix factorization in clustering
- Graph augmented factorization
- Local anomaly detection and implementation in Python
Non-negative Matrix Factorization in clustering (NMF)
Non-negative matrix factorization perform unsupervised clustering by factorization of a matrix $V$ into left and right matrices $W$ and $H$. The right matrix $H$ is interpreted as containing basis vectors for the feature space in $V$ and the rows of the left matrix contain weights for each basis such that $V_{i} \approx W_i\times H$. That is, any row $V_i$ in $V$ is approximated by the weighted combination of basis vectors in $H$. $W$ and $H$ are constrained such that all elements are required to be non-negative.
Let $V$ be a $M\times N$ matrix. $W$ and $H$ are then $M\times P$ and $P\times N$ respectively. The factorization achieves dimensionality reduction (compression) through the $P$ number of latent dimensions.
Selecting $P$ is a problem design choice. Naturally, a larger choice of $P$ allows the decomposition more latent degrees of freedom while a small $P$ puts emphasis on compressing the feature space into fewer latent dimensions.
In generally the problem of computing $V \approx W\times H$ is an optimization problem where the loss is computed as:
Non-negativity contributes to the interpretabilty of the resulting factorization. However, it is assumed that the features of $V$ are or can be made positive which is not always true.
Graph augmented factorization
The paper [1] propose guiding the factorization using a measure of similarity between observations in $V$, towards the goal of detecting outliers. The loss is penalized by similarity matrix $S$. $S$ is created by, first, computing the pairwise distances between observations in $V$ and, second, deriving the shortest path between all observations in a graph traversing sense.
The new loss function is
where $\alpha$ is used to trade-off between the new and original NMF cost function and $\gamma$ control regularization of $H$ and $W$.
Similarity matrix
The similarity matrix, $S$ is constructed in three steps. First the pairwise distances between all observations is computed in $D$:
. Note that the diagonal of $D$ is zero.
Second, $D$ is interpreted as a graph such that an elemnt $D_{ij}$ is interpreted as the cost (or weight) of transitioning from state $i$ to state $j$. Third, the path that traverse all nodes and whose sum of weights is the lowest is computed - this is called the Minimum Spanning Tree (MST).
The example below show the graph representation of three samples in $D$ and its corresponding MST:
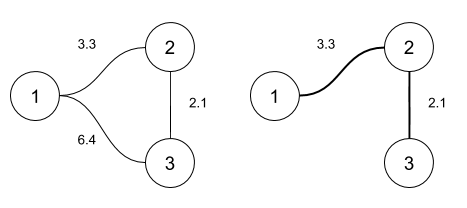
Figure: Matrix interpretation as a graph (left) and its Minimum Spanning Tree (right)
Last, to convert the MST into a similarity matrix, the positive elements of the MST are inverted
Similarity information is incorporated by the term $||S - WW^T||_F$ in the objective function. This way, $W$ is penalized for deviating from $S$.
Local anomaly detection
Having computed the factorization $WH$ from observations $\lbrace v_i \rbrace$ in the training set $V$, a sample $o$ is classified as inlier or outlier by computing the distance between the sample and the cluster it is most similar to:
From this point, the distance $d_o = ||o - w_p||$ is used to rank or discriminate inliers and outliers. The paper uses a ranking score where the samples with top $K$ distances are considered outliers. Using top $K$ samples this way makes $K$ a design parameter and the paper set it equal to the true number of outliers in each considered dataset.
Results on Lymphography dataset
The authors evaluate anomaly detection performance on a most of datasets. In recerating this paper special attention
had to be given to the initial guess $W_0$ and $H_0$. Using random VCol [2] to initialize $W$ and $H$ proved
significant. The implementation is in Python using Numpy and Scipy.
Objective function is:
from numpy.linalg import norm # (frobenius)
# ...
def objective(theta):
# restructure the (flat) hyper-parameters in theta into W and H
W, H = inflate(theta)
err = norm(S - W.dot(W.T))**2 + alpha*norm(V - W.dot(H))**2 + beta*(norm(W)**2 + norm(H)**2)
return err
Note, the paper uses Stochastic Gradient Descent for computing $W$ and $H$ and the objective is slightly different due to that. However, using a black-box minimizer resulted in fairly good performance:
from scipy.optimzie import minimize
# ...
# restructure W and H into one flat array
theta = defalte(W,H)
maxiter = 50
non_negative_bounds = [(0,None) for i in range(len(theta))]
result = minimize(
obj,
theta,
bounds=non_negative_bounds,
method='SLSQP',
tol=1e-9,
options={'disp':False, 'maxiter':maxiter}
)
W, H = inflate(result.x)
which detect 6 out of 6 anomalies in the Lymphography dataset [3] after less than 50 iterations.
References
- I. Ahmed, X. B. Hu, M. P. Acharya, Y. Ding 2020. Neighborhood Struccture Assisted Non-negative Matrix Factorization and its Application in Unsupervised Point Anomaly Detection. arXiv:2001.06431
- R. Albright, C. D. Meyer, A. N. Langville 2006. Algorithms, initializations, and convergence for the nonnegative matrix factorization. NCSU Technical Report Math 81706, NC State University, Releigh.
- M. Zwitter, M. Soklic University Medical Centre, Institute of Oncology, Ljubljana, Yugoslavia 1988 https://archive.ics.uci.edu/ml/datasets/Lymphography